This is a guest blog from Dr Ann Wheeler, Head of the Institute of Genetics and Cancer Advanced Imaging Resource, showcasing their exemplar for metadata capture and data management workflows
When the Edinburgh Bioimaging team, a group of technical specialists and core facility staff, first convened, the question of “What to do with all the data?” was a hot topic. The University of Edinburgh has invested over £50 million in bioimaging equipment, which can generate terabytes of data annually. Managing such large datasets presents significant challenges.
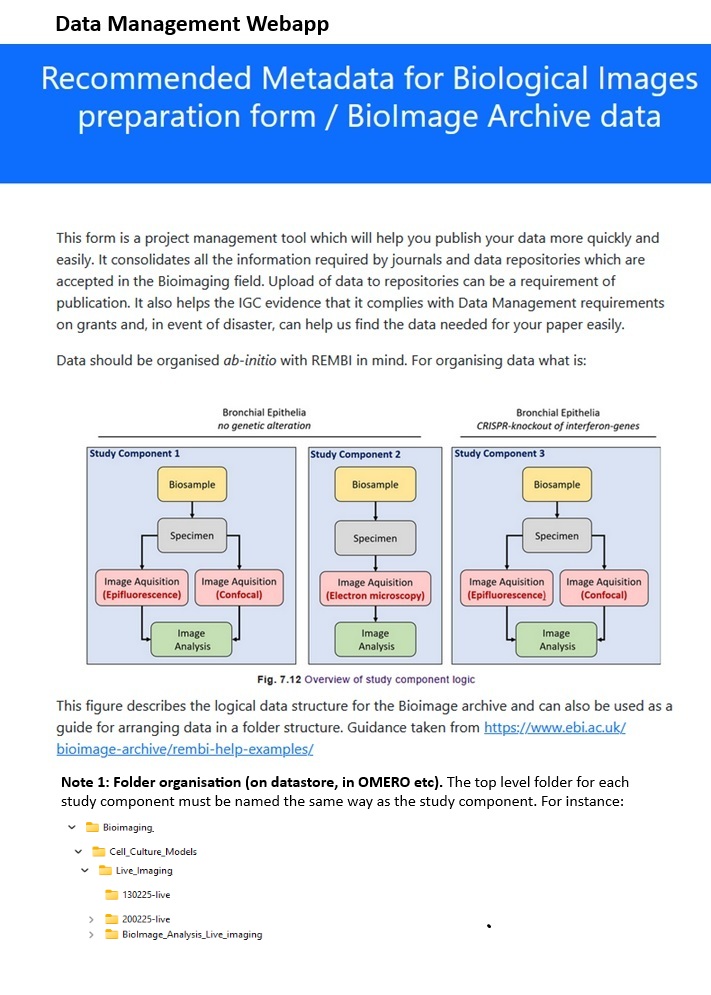
During the COVID period, leaders in the bioimaging field addressed these challenges by publishing metadata recommendations for experiments: REMBI- Recommended Metadata for Biological Images. Additionally, the European Bioinformatics Institute launched the Bioimage Archive, a repository for extensive imaging data, which some journals now require for data submission prior to publication. This is important for those of us who generate, or own bioimaging data to note!
A guide to capturing Recommended Metadata for Biological Images (REMBI)
While this is all great for the wider community, for those of us supporting data generation in Edinburgh, challenges remain:
- Bioimaging core facility staff aren’t the data owners.
- Data storage methods vary widely across departments.
- Using community data repositories like OMERO would require substantial restructuring.
- Re-provisioning costs are high in terms of both staff time and finances.
So, how can we promote data management consistent with FAIR (Fair, Accessible, Interoperable and Reusable) and REMBI (Recommended Metadata for Biological Images) principles at no cost?
Fortunately, collaboration between Bioimaging staff, College of Medicine and Veterinary Medicine (CMVM) data managers, and IT colleagues focused on data management, provided a solution.
We decided on a system involving a form and two tables to record metadata not captured by bioimaging equipment:
- The form: Describes the experiment (e.g., cell types, preparation for imaging).
- Table 1: Details imaging data acquisition methods (e.g., microscope type, live or fixed experiments).
- Table 2: Outlines image data analysis methodology. This is separate as analysis can occur later and may involve different personnel.

Example of data management form.
At its simplest, CMVM data managers can distribute these forms to data owners, who can fill them out and store them with their data. The next step was developing a WebApp for core facility SharePoint sites. This app facilitates easy data entry and sends submissions to the data owner and the relevant CMVM data manager. Submissions can be downloaded and stored alongside the data or within any chosen data management structure. The forms are flexible, allowing for updates or multiple entries if data are analysed in different ways.
The workflow, created in consultation with Bioimage Archive staff, complies with REMBI standards. Completing the forms thoroughly generates metadata, simplifying data uploads to the Bioimage Archive, and compliance with FAIR principle regardless of storage method.
This flexible, no-cost solution meets the needs of all CMVM departments and is potentially beneficial for colleagues in the College of Science and Engineering and the broader community. It’s easy to implement, use, and was well-received by the CMVM community.
