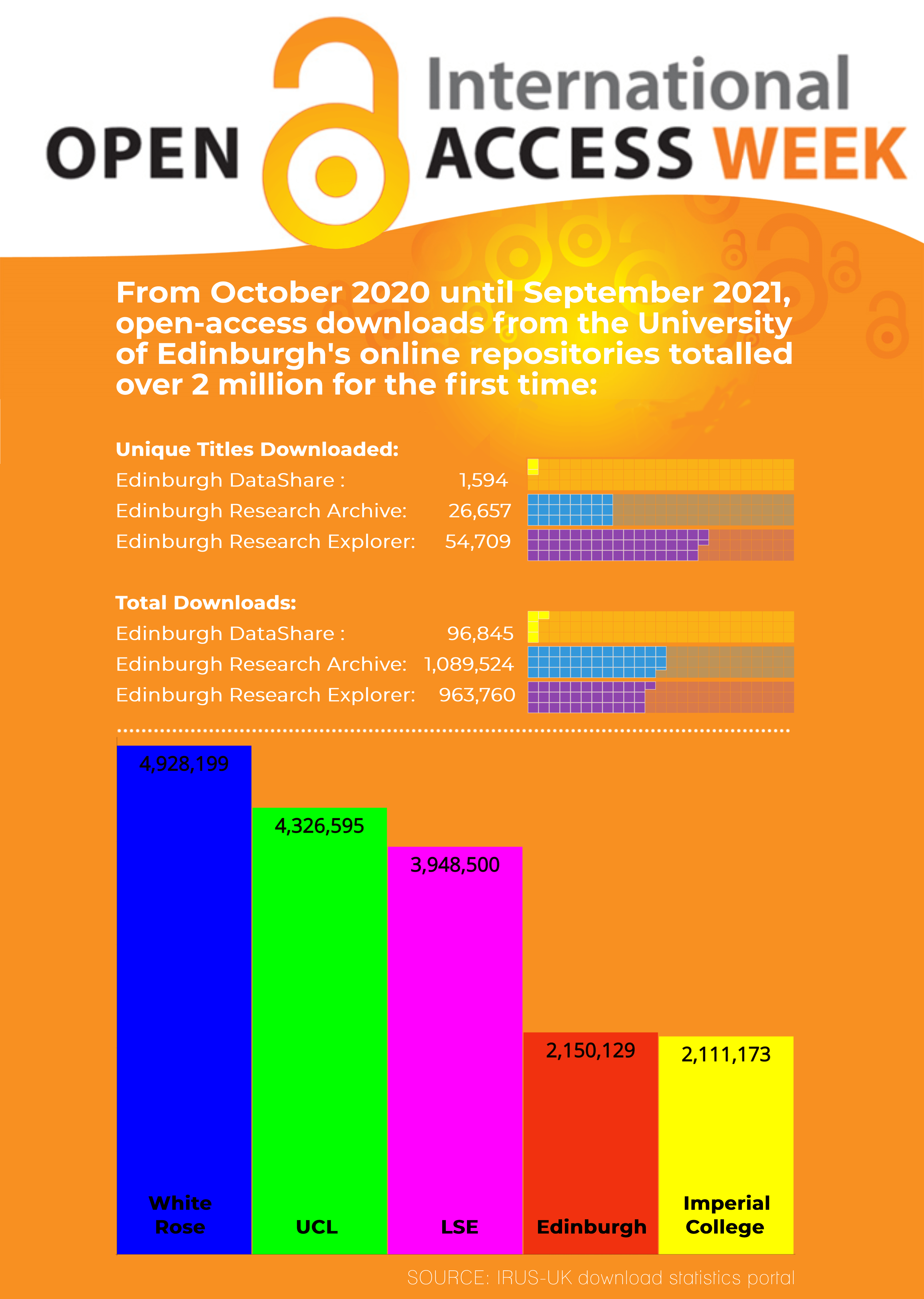
2022 has been a big year for open access at the University of Edinburgh. We started the year off with a bang by introducing a revised Research Publications & Copyright policy in January. This mandatory open access policy applies to all University staff members with a responsibility for research. Going forwards all authors automatically grant the University a non‐exclusive, irrevocable, worldwide licence to make manuscripts of their scholarly articles publicly available under the terms of a Creative Commons Attribution (CC BY) licence.
We are proud that the University of Edinburgh was the first institution in the UK to adopt this type of progressive rights retention open access policy, and we are hugely encouraged to see that many other universities are adopting similar policies. In many ways the policy is just the starting point and most of the hard work actually happens afterwards when supporting members of staff with new publishing processes. This post is intended to give an update of progress so far and to give an account of how the University of Edinburgh policy is having a positive effect on the number of open access publications being immediately available.
Contacting authors
During 2022 the Library in conjunction with Professional Service staff embedded within Schools and Colleges have held an ongoing series of internal seminars for Schools/Institutes. To date the recorded attendance has been 1700 researchers (21.5% of 7,900 total). During these sessions academic staff were informed about the new institutional policy and the support options that are available to them. A second approach was to send All-Staff emails circulated by local College and Schools with information about the new policy and guidelines. This co-ordination between the Library and Professional Service staff was essential in spreading the message about the new institutional policy and getting widespread buy-in from academic staff.
Contacting publishers
In parallel to informing academic staff the University undertook an exercise to formally write to and inform the most popular publishers that University staff submit their work to. Solicitors from our Legal Services department sent a Notice of Grant of Licence by recorded delivery and email to 170 publishers we identified. These publishers we selected covered in the region of 95% of the University’s publication outputs.
Many publishers have introduced restrictive publishing agreements which require embargo periods, and some publishers even assert that their licensing terms will supersede any other prior agreements. We dispute this and if challenged the University will be able to bring a legal claim against the publisher as they have willingly procured a breach of contract against our pre-existing rights.
For a claim of procuring a breach of contract to succeed it must be shown that the defendant knew about the prior contract and intended to encourage another person to break it. Our solicitors have prepared a sworn Affidavit confirming service on all the recipients which will be sufficient to confirm that all the named publishers were indeed advised of our position ahead of article publication and that they have subsequently asked an author to breach the terms of their employment contract by accepting a publishing licence. Most of the time these publishing agreements are click-thru licences that are impossible to edit or change.
With a prior licence granted and publishers contacted the University can now deposit the accepted manuscript of articles in our digital repository, with the article metadata usually available immediately upon deposit and the scholarly article made accessible to the public on the date of first online publication under a Creative Commons Attribution (CC BY) licence.
A snapshot of open access statuses
So with the policy in place, and the majority of authors informed, how effective has the policy been over the first 9 months?
Using data from our Current Research Information System (CRIS) we can track progress on articles published and their open access status. However, we have to be aware of some of the limitations about the health of the metadata extracted from the CRIS. Sadly around 5% of research output records do not have complete metadata allowing us to determine their status. Another point to note is that individual research outputs records in the CRIS commonly have information about both the accepted manuscript version and a final published version. Due to the data model of the CRIS it is not easy to disambiguate between the green versus gold OA statuses when running reports.
Bearing this in mind the table below shows the current open access status of 2022 journal articles:
| Open status |
Count of OA status |
Percentage |
| Closed |
219 |
5% |
| Embargoed |
576 |
12% |
| Indeterminate |
226 |
5% |
| Open |
3519 |
78% |
| Grand Total |
4540 |
100% |
For the 2022 calendar to the present date (mid-Oct) University staff members have written and published 4737 journal articles, of which 197 are not yet published so we’ll discount those. This table includes journal articles that are published via all types of OA route – including fully Gold OA journals and Gold OA via Transformative Agreements (Read & Publish deals). If we remove the journal articles that have been published under a Transformative Agreement or in a fully open access journal we will be left with the number of Green OA articles. We have determined that 958 journal articles have made open access via the repository (Green OA) route in the first 9 months of 2022. It is interesting to note that a Rights Retention Statement (RRS) was only included in 103 articles.
Timing of Open Access
By comparing the dates we have recorded for the official publication date and the date of earliest online access via the repository we can tell something about the timing of open access. The table below shows the available data for articles published in 2022.
| Timing of OA |
# of Green OA articles |
| Before publication |
157 |
| Within 1 month of publication date |
644 |
| Between 1-3 months of publication date |
58 |
| Between 3-6 months of publication date |
16 |
| Greater than 6 months of publication date |
83 |
The majority of articles released before official publication in the journals are preprints that have been deposited in arXiv (or similar) and share a single research output record for the preprint version and journal article.
Around two-thirds (67%) of the Green OA articles were deposited and made OA within 1 month of the date of publication. This time period is the one recommended by UKRI to comply with their 2022 Open Access policy. Around 16% of Green OA articles were made open access after this 1 month deadline so technically would not comply with research funders policies, like the UKRI. Looking at the data more closely reveals that the majority of these seemingly uncompliant articles were submitted before 2022 so actually would not need to follow the immediate OA requirements.
Progress: snapshot of licensing formats
Looking at the licence details on the open access research output records in the CRIS (which includes Gold OA and Transformative Agreements) show that the Creative Commons Attribution (CC-BY) licence is the most popular. It is worth noting that many scholars have opted to choose the more restrictive Creative Commons Attribution-NonCommercial-NoDerivatives (CC BY-NC-ND) licence. The licence field is not mandatory in the CRIS which means that often is it not completed. This is reflected by the high ‘unspecified’ response recorded for 1311 records.
| Row Labels |
Count of Licenses to electronic version documents |
| All Rights Reserved |
56 |
| Creative Commons: Attribution (CC-BY) |
2433 |
| Creative Commons: Attribution No Derivatives (CC-BY-ND) |
10 |
| Creative Commons: Attribution Non-Commercial (CC-BY-NC) |
244 |
| Creative Commons: Attribution-NonCommercial-NoDerivatives (CC BY-NC-ND) |
551 |
| Creative Commons: Attribution-NonCommercial-ShareAlike (CC BY-NC-SA) |
20 |
| Creative Commons: Attribution-ShareAlike (CC-BY-SA) |
1 |
| GNU GPL |
1 |
| Other |
55 |
| Unspecified |
1311 |
| Grand Total |
3519 |
Summary
During the first 9 months of this year professional service staff in conjunction with the Library have been rolling out a massive advocacy push to let authors know about the new research publications policy. This has led to widespread engagement with academic authors who have made 90% of their research outputs open access, mostly within one month of publication. So far we have been informed that only three academics have opted-out from the institutional policy. This relatively low drop-out rate when considered alongside the fact that only 10% of accepted manuscripts have included the rights retention language (RRS) may mean that more outreach activities are required.
We note that the majority of these outputs have been made open via a combination of Transformational Agreements and Gold Open Access journals. However, where this has not been possible, the open access policy has enabled the remaining 27% to be published via the repository Green OA route mostly without embargo. We believe the rights retention policy is an important mechanism to allow authors to publish their research immediately, regardless of whether they have research funders mandates or not.




 Much has been recently been written about the value of preprints which facilitate rapid and open dissemination of research findings to a global audience (if you’d like to read more about the rise of preprints in the life sciences I would recommend this
Much has been recently been written about the value of preprints which facilitate rapid and open dissemination of research findings to a global audience (if you’d like to read more about the rise of preprints in the life sciences I would recommend this